How to order?
To order easily and quickly, you can:
- send us an email at contact@gl-biocontrol.com,
- call us by phone at +33 9 67 39 35 20,
- send us a fax at +33 9 55 25 40 31.
Can I be delivered on Saturday?
Products can be delivered on Saturdays in France. Simply add a comment when you order. For other countries, please contact us:
Phone: + 33 (0)9 67 39 35 20
contact@gl-biocontrol.com
How to pay?
Payement has to be done via bank transfer on GL BIOCONTROL account. Other payement methods can be considered but require prior agreement of GL BIOCONTROL.
What if nobody is available to receive a delivery?
The carrier will leave a delivery notice with its contact information.
What is the delivery time of the products?
We do our best to deliver the products within 48 hours in France.
What to do in case of delivery errors?
Delivery errors can be on the product reference or on the quantity. Please, in either case, contact us as soon as possible. Indeed, if products have to be returned, you have a delay of 8 business days (except products to be kept cold).
ATP AND ATP-METRY
What is ATP?
Adenosine triphosphate (ATP) is the major intermediary energy required in most cellular metabolism reactions. Every living cell produces and consumes ATP. This coenzyme, specific to living environments, proves the existence of living organisms.
Which microorganisms are lysed by the DENDRIDIAG® reagent?
DENDRIDIAG® kits preferentially lyses bacteria, cyanobacteria and amoeba. For total lysis of all microorganisms (fungi, yeast, algae…) consult GL BIOCONTROL.
How and how long can I keep the plastic consumables?
Plastic consumables must be stored in a dry area at room temperature. Their expiration date is displayed on their individual packaging.
What do I measure with ATP-metry?
Quantifying ATP equates to quantifying total microorganisms (or total flora) of a sample. ATP-metry is a biomolecular technique, based on bioluminescence. The measurement is done using a luminometer.
At what temperature should I use the reagents?
To ensure a maximal enzyme efficiency, DENDRIDIAG® reagent and STANDARD 1000 must be used at room temperature (18°C – 25°C).
I forgot the DENDRIDIAG® reagent at room temperature. What should I do?
The kits IW, SW and BF can be stored at room temperature for 3 months.
The kits UPW and AIR must be stored in a freezer (-18°C). If you forgot these reagents at room temperature for a few days, you can perform a control of the reagent efficiency. To do so, refer to the paragraph Control of the reagent efficiency in the handbook’s kit.
What do I measure with the DENDRIDIAG® kits?
With DENDRIDIAG® kits and filtration, only intracellular ATP is measured. It corresponds to the ATP found inside the living cells representative of living bacteria.
Extracellular ATP is also found in sample as a free molecule in the sample. It comes from dead or dying microorganisms. Filtration eliminates free ATP. Without filtration, total ATP is measured: intracellular and extracellular ATP.
How and how long can I store the reagents?
The kits IW, SW and BF can be stored at room temperature for 3 months or in the fridge for over a year.
The kits UPW and AIR must be stored in the dark in a freezer (-18°C). In this way, they can be kept for at least 12 months. After first use, the reagents will be preferentially refrozen. If not, they can be refrigerated (between 3 and 8°C) for a maximum of 8 consecutive weeks.
In which areas of application can use the DENDRIDIAG® kits?
Industrial Water (IW): cooling system, process water circuit, water supply system for industrial purposes…
Sanitary Water (SW): drinking water supply system, water network of spa facilities…
Ultra-Pure Water (UPW): water loop system for medical, pharmaceutical or microelectronics use, water networks under microbiological control…
Surface (BF): swimming pools, food processing, cooling tower, water supply system…
Air (AIR): aeraulic network, hospitals, offices, high risks industries like composting, methanation, farming…
DENDRIDIAG® IW, SW AND UPW KITS
What volume of sample should I filter?
By default, we advise you to filter:
– 10 ml for the IW kit
– 50 ml for the SW kit
– 100 ml for the UPW kit
It is necessary to filter a representative volume of sample in order to get a better reliability of the results.
Anyway, always write down the volume filtered for each sample.
I cannot filter all the sample.
If the sample is highly contaminated, it is possible to clog the filter.
- If you managed to filter at least 10% of the sample: write down the volume filtered, unscrew the filter and empty the syringe. Put the piston back in the syringe placing the black Teflon part at 3 ml if you use a 10 ml syringe or at 10 ml if you use a 50 ml syringe. Screw the filter back on the syringe and follow the classical protocol.
- If you did not manage to filter the sample: pour only 1 ml of the sample in the syringe and complete to 10 ml with sterile water if you use a 10 ml syringe or, pour 5 ml of the sample in the syringe and complete to 50 ml with sterile water if you use a 50 ml syringe.
Consult GL BIOCONTROL for further information.
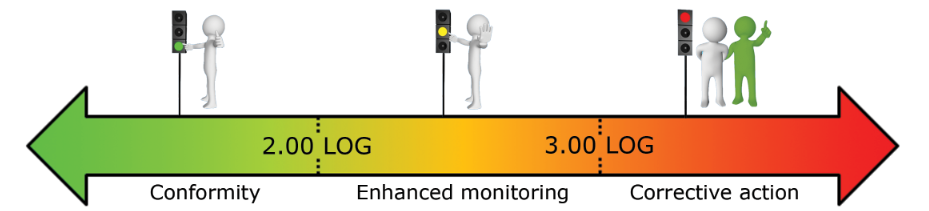
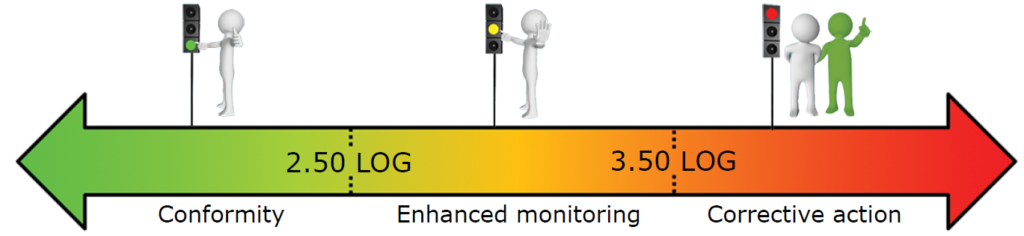
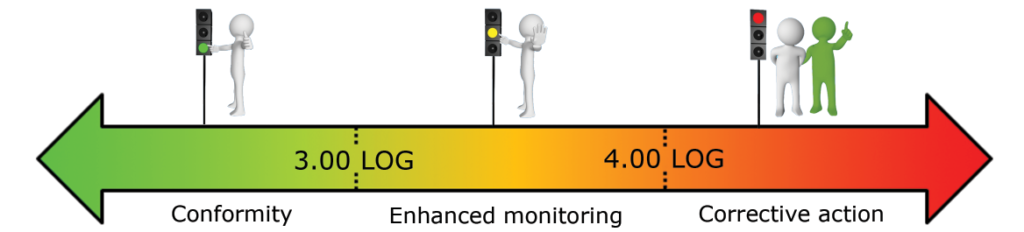
What reference limits should I use for my water analysis?
Based on our experiment, we established the following warning and alarm thresholds:
- Drinking water production system (in LOG eq.bact./ml):
- Drinking water supply network (in LOG eq.bact./ml):
- Domestic hot water – Makeup water (in LOG eq.bact./ml):
- Process water – Cooling tower water (in LOG eq.bact./ml):
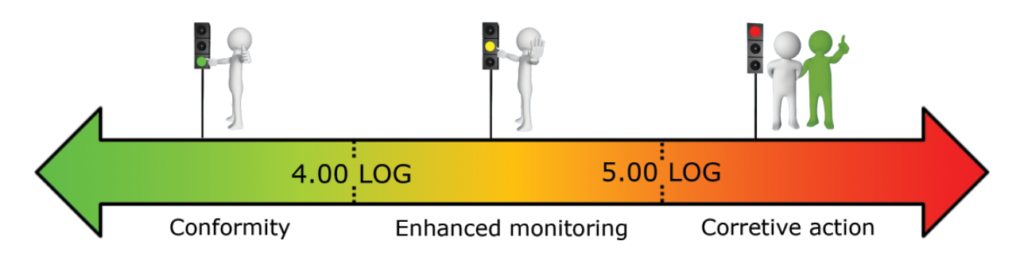
These thresholds should be refined based on the first results obtained on your network.
I cannot get the foam out.
After sucking up the reagent, it is necessary to push all the sample out of the syringe, until a white foam comes out. To get the foam out on a 50 ml syringe, apply a strong and constant pressure of few seconds on the piston with the palm of the hand.
If you continue to have difficulties, we can provide you with 10 ml syringes.
Which unit should I use to express my results?
Picogram ATP per milliliter (pgATP/ml) is the real unit.
For a better understanding of the results, it is possible to use the unit equivalent bacteria per milliliter (eq.bact./ml) using the scientific consensus 1 pgATP ≈ 1 000 bacteria. This result is not rigorously true because the ATP concentration varies from microorganism to microorganism and also differs following the metabolic state of the bacteria.
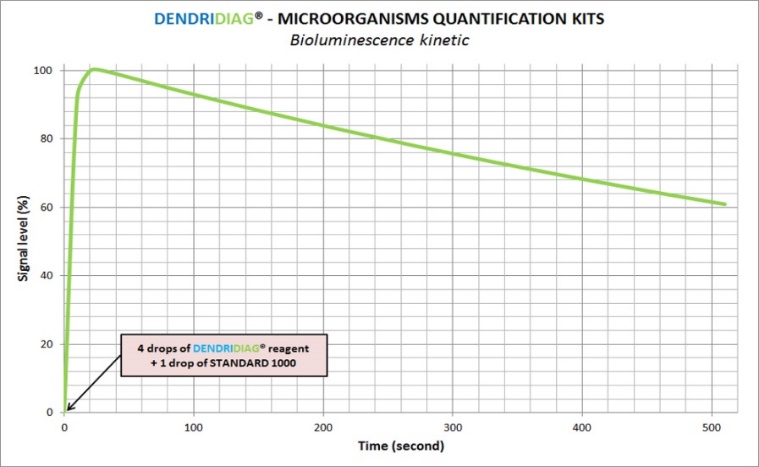
Why is the R2 value inferior to the R1 value?
Signal loss was estimated to 6% to 8% per minute. If measurement takes a long time and there are several minutes between ATP extraction and reading of R2, standardization will happen during the signal degrowth phase. It is often observed on sample highly contaminated.
We advise you to restart the analysis by filtrating a smaller volume of water.
Too much foam in the test tube.
To recover all the microorganisms in the test tube, it is necessary to push all the sample out of the syringe, until a white foam comes out. However, the pressure on the piston must be stopped as soon as the foam comes out in order to avoid formation of a “plug” between the top and the bottom of the tube. During manipulation, lean the test tube so the reagent runs along the tube wall.
If you have an excess of foam or a bubble on the upper part of the test tube, tap the bottom of the tube on a flat surface.
Why express my results in LOG?
Results are often expressed in LOG to simplify the interpretation. Indeed, we consider that two results are significantly different if there is a difference of 1 LOG between the two values.
The following table shows you the conversion of eq.bact./ml in LOG:
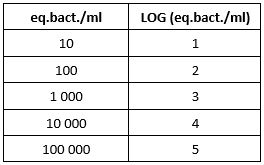
The Excel table and the WebApp DENDRIDIAG®SOFTWARE automatically give you this value.
DENDRIDIAG® BF KIT
What surface dimension should I sample?
Our kit is supplied with a sampling template which defines a surface of 20 cm². With this sampling template, you get a good reproducibility and a significant sampling of the surface studied which leads to reliable results. Nevertheless, if you want to increase the measurement sensitivity, you can perform two sampling of the same surface by moving the sampling template.
Always write down the surface sampled.
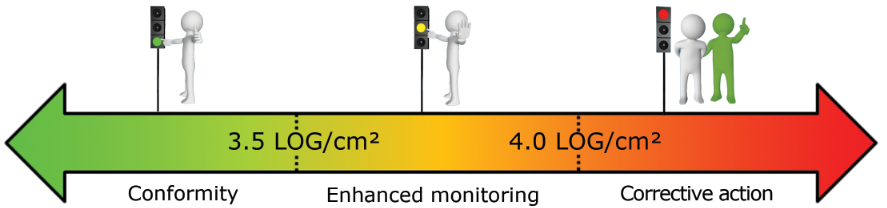
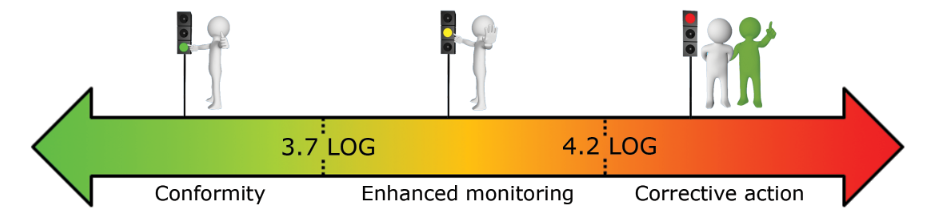
What reference limits should I use for my surface analysis?
Based on our experiment, we established the following warning and alarm thresholds:
- Pool opening (in LOG/cm²):
- Operating swimming-pools (in LOG/cm²):
- Surfaces of water pipes (in pgATP/cm²):
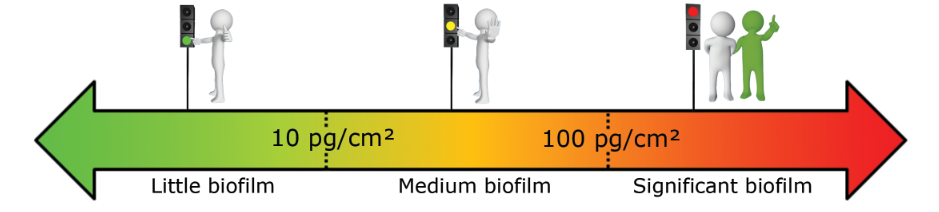
These thresholds should be refined based on the first results obtained on your network.
Which unit should I use to express my results?
Picogram ATP per square centimeter (pgATP/cm²) is the real unit.
For a better understanding of the results, it is possible to use the unit equivalent bacteria per square centimeter (eq.bact./cm²) using the scientific consensus 1 pgATP ≈ 1 000 bacteria. This result is not rigorously true because the ATP concentration varies from microorganism to microorganism and also differs following the metabolic state of the bacteria.
Why express my results in LOG?
Results are often expressed in LOG to simplify the interpretation. Indeed, we consider that two results are significantly different if there is a difference of 1 LOG between the two values.
The following table shows you the conversion of eq.bact./cm² in LOG:
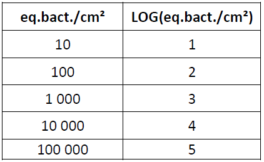
The Excel table automatically gives you this value.
LUMINOMETER
My tube holder for SMART luminometer is dirty. How to clean it?
An excess of foam or a bubble in the upper part of the tube can dirty the tube holder. In this case, you can use a paper towel impregnated with alcohol 70% of water to clean the tube holder.
The luminometer C110 displays « OVERSCALE ».
Luminometer C110 is very sensitive. The message “OVERSCALE” is displayed when the sample is strongly contaminated and the luminometer cannot measure the RLU. If you analyze liquids, restart the measurement by filtrating 1/10th of the volume.
I forgot to write down the value of R1 and R2.
It is possible to retrieve the results in RLU in the luminometer. To do so, turn on the luminometer and after the 10 seconds of calibration, press the up arrow to get the last values obtained.
Luminometer SMART rings and does not complete calibration.
The calibration is done when the luminometer is empty and the cap correctly closed. If the luminometer rings, make sure the tube holder is out and the cap is well closed.